
MEDSUM Tutorial page
Although MEDSUM is very simple to use, there are plenty of tips and tricks that will make your use of MEDSUM more flexible and powerful.
Profiling authors and journals is fun, but MEDSUM is also designed for much more serious research-on-research capabilities for writing
review articles or grant applications. This tutorial guides through the basic use and purpose of the four buttons, with examples
and explanations of their additional capacities:
 Quick Call
Quick Call
The purpose of the Quick Call button is simply to tell you:
1) how PubMed interprets what you just wrote and
2) how many records there are associated with that query.
The query should normally take under 2 seconds to process.
Why use Quick Call? In a standard PubMed search, it may not be so critical to get your search term 100% accurate if you are just looking for
interesting papers, but if you are using MEDSUM to profile fields of research for the sake of a review paper, then you need to check that your
search term is being interpreted correctly.
 EXAMPLE: Try putting in major depression then click Quick Call... you get:
EXAMPLE: Try putting in major depression then click Quick Call... you get:
Note that Pubmed translates your query into: "major depressive disorder"[Text Word] OR "depressive disorder, major"[MeSH Terms]
OR major depression[Text Word]
 TIP: If you think that any of these sub-terms is really what you want, you can just highlight that text,
copy it and paste it into the search box.
TIP: If you think that any of these sub-terms is really what you want, you can just highlight that text,
copy it and paste it into the search box.
 TIP: If you don't like how PubMed splinters your query phrase into various different queries and want to
search purely on the phrase you put in, just put it in double quotes. E.g. compare the Quick Call results of general cognitive ability with
"general cognitive ability" (which is a term often used synonymously with IQ).
TIP: If you don't like how PubMed splinters your query phrase into various different queries and want to
search purely on the phrase you put in, just put it in double quotes. E.g. compare the Quick Call results of general cognitive ability with
"general cognitive ability" (which is a term often used synonymously with IQ).
(Note that in PubMed you can see the interpretation of your search term by clicking on the "Details" tab beneath the search bar.)
 Timeline
Timeline
The Timeline button takes your query and first counts all associated records (publications) between 1945 and the current year. It then
breaks down that total count year by year, starting with the current year and working backwards. Timeline stops when it runs out of records
or when it hits 1945. Each year should take as long as a Quick Call - i.e. between 0 and 2 seconds.
Although the PROFILE button will also give you a breakdown over years as part of its information set, Timeline is much faster for larger number of
records (approx > 1,000 or 2,000) and not limited by number of records (PROFILE is currently limited to 10,000 records).
 EXAMPLE: To demonstrate the speed by which Timeline can summarise millions of records, put mice in the search box and then
click Timeline. You will see approximately 0.8 million records being sorted by year. If you then put rats in the search box and click
Timeline again, you will see the 1.2 million papers associated with rats being broken down by year. Comparing the two sets of data will show
you that papers on mice overtook papers on rats somewhere around 2001/2002.
EXAMPLE: To demonstrate the speed by which Timeline can summarise millions of records, put mice in the search box and then
click Timeline. You will see approximately 0.8 million records being sorted by year. If you then put rats in the search box and click
Timeline again, you will see the 1.2 million papers associated with rats being broken down by year. Comparing the two sets of data will show
you that papers on mice overtook papers on rats somewhere around 2001/2002.
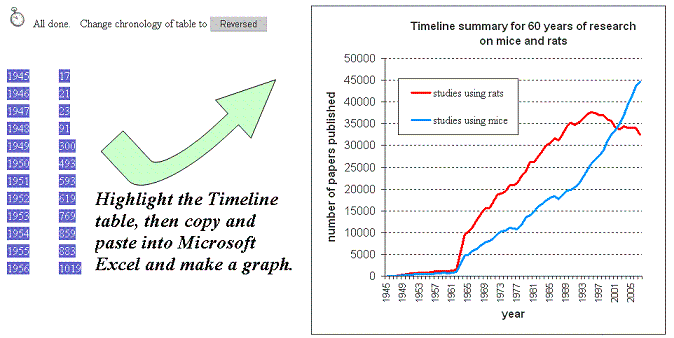
 TIP: Limiting the time span in Timeline...
TIP: Limiting the time span in Timeline...
Often you will not be interested in the older literature on common terms, but rather just want to see data concerning current trends. To
stop Timeline from wasting its time and yours by going back too far, just limit the years using the Date Published [dp] search tag. For
example, to look at papers on schizophrenia from 1980 onwards, type Schizophrenia 1980:2100[dp] then click Timeline. It now
stops at 1980. I use the year 2100, but you can use any random future year.
 TIP: Finding original sources...
TIP: Finding original sources...
You can use finished Timeline results to get an original citation very directly. An example: Enter
Long-Term Potentiation in the search box and click Timeline. Timeline runs out of records in the year 1973. If you then go to
PubMed and enter Long-Term Potentiation 1973[dp], you will get the Bliss & Lomo (1973) paper which is the first publication on
long-term potentiation. Similarly if you enter DRD4 in the search box and click Timeline, the records run back to the year 1991
where there were 3 papers. Typing DRD4 1991[dp] in PubMed gives those 3 papers and it quickly becomes apparent that the Van Tol et al.
April paper in Nature is the original characterisation of the receptor. You can also, of course, find original citations directly in PubMed.
After any search, just select Sort By "Pub Date", then enter the total number of pages in the Page box (i.e. get to the last page), and scroll
to the bottom. I just thought it worth mentioning as I know too many people paper-chase through references to try and find an original
source!
 WARNING: The [dp] search tag...
WARNING: The [dp] search tag...
If you "check" the results of a Timeline (or PROFILE) year against PubMed (or MEDSUM Quick Count), you may be horrified to see that
numbers do not match for recent years. For example, with the term EEG, Timeline or PROFILE will tell you that the year 2004 had 3230
papers on EEG. However, if you put EEG 2004[dp] into PubMed or into MEDSUM Quick Count, they will both tell you that there were 3383
papers. Which is correct? Actually, it's Timeline and PROFILE. The problem is that the [dp] search tag looks for papers that were print
published OR e-published in that year. For papers that are e-published in one year and print published in the next... you can see the problem. And this
is an increasing trend. With the EEG example (in PubMed), if you go to Sort By and choose Pub Date, you'll see the first paper was e-published
in 2004, but finally published in 2006! Thereafter there are 152 papers that were e-published in 2004 and print published in 2005. This
explains the difference of 153 papers between Timeline/PROFILE results and PubMed [dp] results. Timeline and PROFILE use only the most recent
date (i.e. print published, unless it is still only e-published) so that each paper is allocated to only one year. This makes the total number
of papers correct. NOTE: this is also 100% consistent with the DP- fields which are the official "date published" tags associated with each
paper, as can be seen by selecting Display --> MEDLINE in PubMed. It is a problem that the [dp] search tags do not map onto the DP- field
tags... but note that PubMed is primarily designed for finding papers rather than accurately counting numbers of papers, which is what we
want here.
 MeSH It
MeSH It
The purpose of the MeSH It button is to take your query and convert it into Major Medical Subject Heading (MeSH) terms. To do this, it
calls up and downloads the most recent 500 papers coded into MEDLINE (i.e. with MeSH terms) and counts up the occurrences of each Major
MeSH heading. It then uses these top 10 terms as a seed list to get the real frequencies of the terms (in conjunction with your query) in
the entire MEDLINE database. It then returns these to you. MeSH It usually takes between 20 and 60 seconds to run.
What are MeSH terms? If you go to PubMed and run a query on Epilepsy 2001[dp], then change the Display box (under the search box)
from "Summary" to "MEDLINE", you will see the basic data MeSH It works on. On the first paper, the third field down reads "STAT - MEDLINE"
and this means that the paper has been entered into MEDLINE. In other words, it has been read and coded in terms of standardised medical
terms. You can see these actual terms further down the fields list, as a string of MeSH Headings (MH- ) fields. This explains the MeSH It
icon of the magnifying glass looking at clusters of MH- fields.
The importance of MeSH Headings is that they are very reliable descriptors of papers. A paper with "epilepsy" in the abstract may indeed be
about epilepsy... or it may be a very tentative link with the abstract containing a sentence like "So this new gene expressed in the brain of
D. melanogaster may potentially somehow have some relevance to behavioural, neurological or psychiatric disorders such as depression,
epilepsy, schizophrenia, dementia, bipolar disorder or even extroversion".
Furthermore, MeSH Headings are particularly useful for bioinformatics tools such as BITOLA (see
IBMI webtools homepage). BITOLA links through diverse papers using MeSH terms because MeSH
terms are not only robust descriptors of papers, but also because there are a limited set of MeSH terms. The original purpose of MEDSUM was
simply to be a "free-text to MeSH converter" for BITOLA, but then MEDSUM expanded and the "free-text to MeSH conversion" got relegated to
this MeSH It button. The MeSH It facility is important for BITOLA because many scientists are unfamiliar with even the MeSH terms of their
own publications. More importantly, the correct MeSH term may not be obvious or may be hard to find. For example, if you put "depression"
in BITOLA, hoping to get "major depression" as an option, you wouldn't. However, "depressive" would return the correct options of "Depressive
Disorder" or "Major Depressive Disorder". This particular misunderstanding is easily solved using Search: MeSH on the PubMed site (before the
search box, change Search: PubMed to Search: MeSH), or even by looking at a Quick Call result. But there are more tricky examples...
 EXAMPLE: Consider the case of wanting to use BITOLA to find genes or drugs that have been associated with performance in the Morris water maze, or at
least something similar (i.e. puroportedly measuring spatial navigation capability). Try Morris water maze in Quick Call (fortunately, there
are no papers associated with "Morris AND hydrotherapy AND maze"! so the translation is safe) and then try Morris water maze in Search:
MeSH (Suggestions: "Microbiology, water", "Mocassins, water"??). So those two are no help. However, now put Morris water maze in the
MEDSUM search box and click MeSH It. Simply by taking the first 1000 MEDLINE papers with Morris AND water AND maze in the text, MeSH It can
tell you that over 80% of the time, these papers are classified with the "Maze Learning" Major MeSH Heading... so "Maze Learning" is the
BITOLA term that you probably want.
EXAMPLE: Consider the case of wanting to use BITOLA to find genes or drugs that have been associated with performance in the Morris water maze, or at
least something similar (i.e. puroportedly measuring spatial navigation capability). Try Morris water maze in Quick Call (fortunately, there
are no papers associated with "Morris AND hydrotherapy AND maze"! so the translation is safe) and then try Morris water maze in Search:
MeSH (Suggestions: "Microbiology, water", "Mocassins, water"??). So those two are no help. However, now put Morris water maze in the
MEDSUM search box and click MeSH It. Simply by taking the first 1000 MEDLINE papers with Morris AND water AND maze in the text, MeSH It can
tell you that over 80% of the time, these papers are classified with the "Maze Learning" Major MeSH Heading... so "Maze Learning" is the
BITOLA term that you probably want.
 PROFILE
PROFILE
The PROFILE button is often the most fun of the four MEDSUM buttons. Its most common use is for exploring the publication
history and biases of an individual researcher or a journal. However, it can also be used to explore subject areas very thoroughly. Each
of these areas; authors, journals and subjects have their own tips and tricks associated with them, so they will be dealt with separately.
Very generally, PROFILE downloads the full MEDLINE versions of all papers called by the query (title, abstract, authors, MeSH terms,
everything). As PubMed E-Utilities requests that you download a maximum of 500 records at a time with 3-second delays between each batch,
the PROFILE button currently limits itself to the first 10,000 records (previously was 1,500). This easily covers all author profiles,
all journal profiles, but only some subject profiles.
A PROFILE can take between 2 seconds and a number of minutes, depending on the number of records being downloaded and processed.
 PROFILING an AUTHOR.
PROFILING an AUTHOR.
This is usually relatively quick. PROFILE will present a series of summary statistics.
These are:
Top 10 Authors (or in this case the author in [square brackets] and his/her co-authors, unless there is name ambiguity - see below)
Top 10 Journals (where that author has published)
Humans:Animals ratio (how many papers they have published on humans, and how many on animals)
Male:Female ratio (same as above, but papers on both males and females get one count on each)
Age profile (for studies on humans, the ages of the populations used)
Top 10 Major MeSH Headings (the broad categories of their papers)
Top 10 Starred MeSH Headings (the specific categories and sub-categories of their papers)
Papers sorted by years (same as Timeline - total number of papers tabularised by year)
 TIP: Name ambiguity...
TIP: Name ambiguity...
Middle names are rather useless things until you become a scientist. If your name really is John Smith and you have no middle initial,
a search on Smith J will not be as rewarding as if you have a literature-unique name. (In fact Smith J returns over
15,000 papers). In order to easily write a query that returns your own papers only and gives a representative profile, just utilise your
regular co-authors (e.g. PhD and post-doc supervisors) in the format: Smith J AND (Flanders N OR Burns CM OR Smithers W OR
Terwilliger RU). Although this might not cover all your papers (e.g. sole author papers), nevertheless it will make a decent profile
of your publication history.
 PROFILING a JOURNAL.
PROFILING a JOURNAL.
Journal profiles look identical to author profiles. The only differences are 1) the "Top 10 Authors" are the top 10 publishers in that journal,
and 2) The "Top 10 Journals" are usually only the journal name in [square brackets] to indicate that it recognised that it was a journal profile.
Otherwise, the Humans:Animals ratio and Major/Starred MeSH Headings give you a good idea if your work fits the journal's areas of interest and/or
readership. The publications by year give an indication of how old the journal is and whether it is growing in size.
 TIP: If you are worried that your profile of a long-established journal is confounded by data from the 70s and 80s and might not represent current
trends in the journal, simply limit your search to current years. For example, with the Journal Behavior Genetics (with PubMed records beginning in
1970, according to Timeline), enter Behavior Genetics 2000:2100[dp] or you can use the format: Behav Genet "last 5 years"[dp].
TIP: If you are worried that your profile of a long-established journal is confounded by data from the 70s and 80s and might not represent current
trends in the journal, simply limit your search to current years. For example, with the Journal Behavior Genetics (with PubMed records beginning in
1970, according to Timeline), enter Behavior Genetics 2000:2100[dp] or you can use the format: Behav Genet "last 5 years"[dp].
 PROFILING a SUBJECT.
PROFILING a SUBJECT.
At the moment, the utility of the PROFILE button to handle subject areas is limited due to the 1500 record limit which is often far too small for
characterizing large domains of research. However, if just looking to characterise recent trends, PROFILE can help identify
the top researchers and journals for the subject, or (via the starred MeSH terms) indicating newer areas of interest associated with the subject.
 TIP: Given that several sets of information (the research ratios, age profiles and Major/Starred MeSH terms) all
come from the MeSH fields, you may want to ensure that the records you process are MEDLINE entered. For subjects that have huge numbers of publications
each year, a SUBJECT PROFILE might only just catch the current year and most of those papers will not yet be MEDLINE entered. For example, enter
magnetic resonance imaging and click PROFILE (chose 1500 records). Look at how many of those records are in MEDLINE. Now try magnetic
resonance imaging AND medline[sb]. You will see that because you are selecting the 1500 most recent papers that also have full MEDLINE entries,
all papers contribute to all descriptive fields.
TIP: Given that several sets of information (the research ratios, age profiles and Major/Starred MeSH terms) all
come from the MeSH fields, you may want to ensure that the records you process are MEDLINE entered. For subjects that have huge numbers of publications
each year, a SUBJECT PROFILE might only just catch the current year and most of those papers will not yet be MEDLINE entered. For example, enter
magnetic resonance imaging and click PROFILE (chose 1500 records). Look at how many of those records are in MEDLINE. Now try magnetic
resonance imaging AND medline[sb]. You will see that because you are selecting the 1500 most recent papers that also have full MEDLINE entries,
all papers contribute to all descriptive fields.
 TIP: Handling the information in chunks...
TIP: Handling the information in chunks...
If you are determined to process all records from a large subject area (with over 10,000 records), you can simply do PROFILE in manageable chunks.
First run your query through Timeline to see how many publications there are for each year, then divide those into spans that fit under 10,000 records.
Years can be chunked in the format: autism 1990:1995[dp]
 WARNING: The [dp] search tag...
WARNING: The [dp] search tag...